Re: GSoC project: K-medoids clustering in Madlib
| From: | Maxence AHLOUCHE <maxence(dot)ahlouche(at)gmail(dot)com> |
|---|---|
| To: | Atri Sharma <atri(dot)jiit(at)gmail(dot)com> |
| Cc: | "Iyer, Rahul" <Rahul(dot)Iyer(at)emc(dot)com>, "pgsql-students(at)postgresql(dot)org" <pgsql-students(at)postgresql(dot)org>, "devel(at)madlib(dot)net" <devel(at)madlib(dot)net>, "Philip, Sujit" <Sujit(dot)Philip(at)emc(dot)com> |
| Subject: | Re: GSoC project: K-medoids clustering in Madlib |
| Date: | 2013-04-21 17:46:46 |
| Message-ID: | CAJeaomW50Bt-RFZs73b6M4iDaqj8mFFrfjXzHsdhSJb4iJzOHQ@mail.gmail.com |
| Views: | Whole Thread | Raw Message | Download mbox | Resend email |
| Thread: | |
| Lists: | pgsql-students |
Hello!
I've pretty much improved my visualizer:
- It is now able to color the points according to their assigned
cluster. It has occured to me last night that it was the main inconvenient
of the k-means algorithm: a point is assigned to the nearest centroid's
cluster, which is easy to compute.
- It's now able to load data from a file, if it has been previously
generated, so it is no longer mandatory to specify the number of clusters
wished when using "old" data.
- It displays the original clustering and the k-means++ clustering for
comparison purposes, and it is very easy to add new clusterings.
- It is available on GitHub!
https://github.com/viodlen/clustering_visualizer
Still, there is room for improvements:
- It only tests with 2-dimensional spaces. This won't evolve, as it
would get difficult to visualize.
- It only uses the euclidean distance. This can be fixed, but would be
heavy to implement, and probably ugly (a hashmap to match the python's
distance function with the MADlib's one).
- For now, the points are reparted in the clusters folowing a gaussian
law (not sure of my vocabulary here). This can be easily fixed, and will
probably be in a future version.
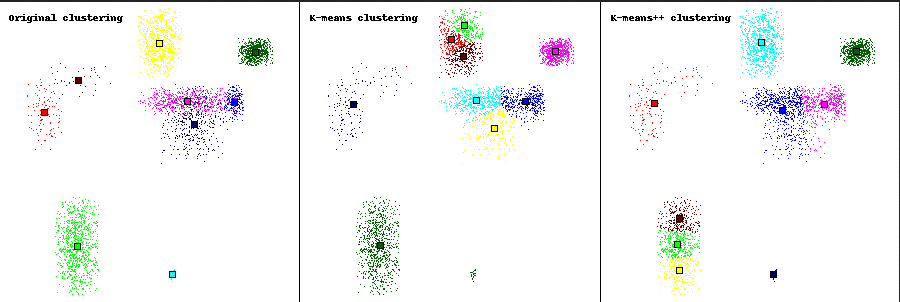
In attachment is an output example. It shows the poor results of the
k-means algorithm on contiguous clusters.
It also shows the interest of the k-means++ algorithm: the small light-blue
cluster (in the original clustering) is correctly identified as a complete
cluster by the k-means++ algorithm, when it was only a part of a bigger
cluster with the simple k-means algorithm.
The characteristics of the k-means algorithm make it easy to calculate the
clusters only from the points and the centroids, but this won't be true for
the k-medoids algorithm. So, sadly, if I implement the latter, it won't be
possible to keep the same function signature: some more data will have to
be returned, along with the centroids list.
I've also wondered if it would be useful to implement the clustering
algorithms for non-float vectors (for example, strings), provided the user
gives a distance function for this type?
--
Maxence Ahlouche
06 06 66 97 00
93 avenue Paul DOUMER
24100 Bergerac
| Attachment | Content-Type | Size |
|---|---|---|
|
image/png | 18.3 KB |
In response to
- Re: GSoC project: K-medoids clustering in Madlib at 2013-04-20 14:59:41 from Atri Sharma
Responses
- Re: GSoC project: K-medoids clustering in Madlib at 2013-04-21 18:03:07 from Atri Sharma
- Re: [MADlib-devel] GSoC project: K-medoids clustering in Madlib at 2013-04-25 05:16:04 from Atri Sharma
Browse pgsql-students by date
| From | Date | Subject | |
|---|---|---|---|
| Next Message | Atri Sharma | 2013-04-21 18:03:07 | Re: GSoC project: K-medoids clustering in Madlib |
| Previous Message | Atri Sharma | 2013-04-20 14:59:41 | Re: GSoC project: K-medoids clustering in Madlib |